Connectomics is the area of neuroscience that aims to collect and curate the entirety of the connections made by all neurons in a brain (the product being called a “connectome”). For the human brain, that would be a data set of 100 billion neurons, each of which is estimated to make 1000-10000 synapses with other neurons (on the order of 1017 connections). The roll-up-your-sleeves-this-will-get-really-messy way of collecting that kind of data is to slice the brain into nanometers-thick sections and to image each slice with an electron-microscope, which has resolution below the nanometer range, and can reveal the structure of cells on a fine scale. In the image from Kristen Harris's lab below you can see a part of a neuron’s dendrite making a synapse with an axon filled with neurotransmitter vesicles; EM images however cannot show individual proteins or molecules).
The challenge is not just in collecting this type of data - actually, some scientists are working to develop high-throughput machines that largely automate the process - but in storing and analyzing it. The storage problem is important but nothing to write home about; ways to store the petabytes upon petabytes should surely become available soon (data center at CERN already processes about one petabyte of data everyday). The more menacing problem is data analysis.
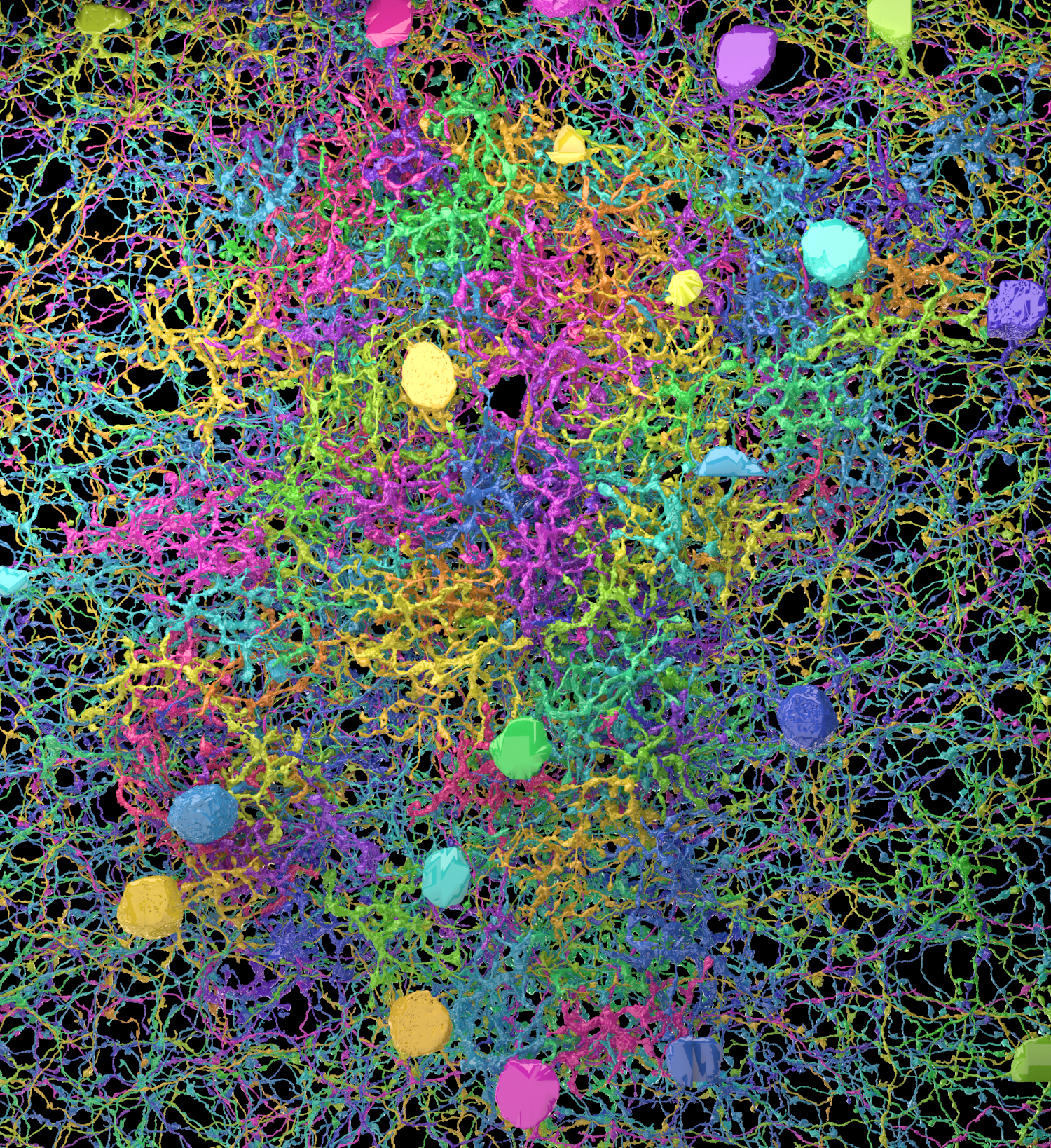
Where does analysis even start? We have thousands of brain slices, each a rain forest of stuff with no visible empty space. The brain is packed with stuff, but unlike a hoarder’s apartment, it’s filled with functional things (even the glia, long derided as “mere support cells” turn out to have serious functional roles). There is no junk in the brain. (In some flyby videos by Terry Sejnowski and Winfried Denk, the viewpoint is from sewer-like tunnels that were once blood vessels). The first order of business is to label every structure in a slice, thus assigning dendrites, axons and cell bodies to different neurons; then, one takes the next slice and labels parts that look like they are contiguous with parts in the previous slice. Stacking together hundreds of these slices, you have what’s starting to look like a neuron. Only about a hundred billion more to go!
Asking undergraduates eager to get some exposure to science to label these images amounts at best to premature squashing of any scientific interests or dreams, and at worst to cruel and unusual punishment. And while computer algorithms are getting better and better at segmenting images, they still lag behind humans in following twisting axons across slices (as one neuroscientist put it, successfully labeling connectomics data pretty much requires us to “solve vision,” referring to the decades-old scientific quest to understand how the brain recognizes objects). This is why Sebastian Seung’s solution is so elegant: turn the labeling into a game and ask thousands of people around the world to play. The EyeWirers, as Sueng called them, not only help to solve scientific problems but get to feel excited by science because they make a direct contribute to it.
Some scientists aren’t too jazzed about connectomics because they feel the ambition of its projects outweighs the possible scientific value. The typical cautionary tale is from the roundworm C. elegans, whose 302 neurons and their connections have been mapped out and yet the complete wiring diagram can generate wildly different patterns of activity. What use is a wiring diagram, if it doesn’t dramatically constrain the possible underlying patterns of activity?
This particular paper demonstrates why connectomics might be useful after all. The problem at hand concerns direction selectivity in the retina. While photoreceptors respond to flashes of light, some downstream neurons within the retina have been found to respond selectively to motion of light in a given direction. Previous experiments have narrowed down the search for direction selectivity to the synapse between bipolar cells (BCs, of which there are five types) and starburst amacrine cells (SACs), but despite various models, the mechanism for how direction selectivity arises hasn’t been found.
Kim et al note data from previous experiments showing that different types of bipolar cells respond to visual stimuli with different temporal dynamics. The BC2 cells respond to visual stimuli about 50-100ms after BC3a, and BC3a responds transiently to changes in light while BC2 responds in a sustained fashion. If a BC2 and BC3a cell were to be active at the same time, their input to the SAC would be stronger (and likelier to produce a strong response in the SAC) than if their activity were asynchronous. Given the time lag between their responses to light, co-activation of a BC2 and BC3a cell would occur if the light hit the BC2 first. One way for these features to create direction selectivity is that the BC2 and BC3a would have to make contacts with the SAC at different parts of its dendritic tree. The work of the EyeWire community shows that this is indeed the case: BC2s tend to contact SACs near the soma, whereas BC3a contact SACs far from the soma.
The authors created a mathematical model to describe direction selectivity in SACs as a result of differential wiring and responses to light in BC2 and BC3a. Similar models have been proposed before (e.g. Barlow and Levick in 1965), but relied on different mechanisms to achieve direction selectivity. The current model combines known responses to light stimulation in different BCs with new data on connection statistics between BCs and SACs. This demonstrates a good use of connectomics data while building a model that makes testable assumptions (e.g. given that BC2 and BC3a respond to light flashes individually as described and that they connect to SACs differentially, would their activities during presentation of moving stimulus follow the predicted time course? And does stimulating BC2 and BC3a at different times relative to each other change how the SAC responds? Do the temporal dynamics of BC2a and BC3 constrain direction selectivity in the SAC?). Rather than being an unattainable or uninterpretable data set, perhaps connectomics will be useful for testing specific hypotheses about wiring in small circuits, and to rule out models that rely on some hypothetical wiring that might not exist. This way, a brain connectome is a start to understanding of neural circuits rather than the end - a necessary but not sufficient framework.
Read more on the EyeWire blog:
http://blog.eyewire.org/eyewires-first-scientific-discovery-and-nature-paper/
And the original paper:
http://www.nature.com/nature/journal/v509/n7500/full/nature13240.html